Refactoring R’s dendrapply
Published:
As someone who specializes in comparative phylogenomics, I work a lot with phylogenetic trees. Trees are represented in R as dendrogram objects, which are essentially a series of nested lists. Each “node” of the tree is a list with multiple members (two if a binary tree, but dendrogram objects are not constrained to be binary), each of which is another dendrogram object. The leaves are special cases in that they have length 1 and an additional property leaf, which is set to TRUE.
R has a number of functions called apply functions, whose primary purpose is to apply a function to a set of things in a particular way. Commonly used examples are lapply to apply a function over a list-like object, tapply to apply a function to a set of objects grouped based on a factor, rapply to recursively apply a function to an object, or apply to apply a function to a matrix/array.
dendrapply is a special type of apply statement intended specifically for applying functions to dendrogram objects. The main requirement is to recursively apply a function to each node of the tree. While this may sound like a task for rapply, rapply is more intended to apply a function to non-list elements of a nested list, while dendrapply is intended to apply the function to all nodes in the list. This means that dendrapply specializes in applying the function to internal nodes of the tree, whereas rapply applies the function only to the leaves (and doesn’t always preserve the original structure).
dendrapply is currently implemented recursively, which has led to some users experiencing issues from stack overflows resulting from deep recursion on trees with many nodes (at least in users’ reports on DECIPHER). Additionally, the recursive implementation makes the function slow for trees with many internal nodes. rapply avoids this issue by factoring out the recursion internally, which gave me the idea to try to implement an optimized version of dendrapply.
Implementation
My focus for this implementation was removing the recursive calls in dendrapply. The final implementation is only a modest runtime improvement, but this can hopefully be optimized prior to the final release.
The full implementation is written in C, which makes a lot of things easier at the cost of a few things becoming really hard. The algorithm proceeds as follows:
copytree = copy(input_dend)
initialize LinkedList
add root node to LinkedList
ptr = head(LinkedList)
while ptr is not NULL:
if node has children:
for child in ptr.node.children:
insert child in next position
else:
while(node has no unevaluated children):
apply function to node
merge node into parent
node = parent
ptr = parent.ptr
ptr = ptr.next
return head(LinkedList).node
It’s a little hard to write out, so I’ll use a small example to showcase. Imagine we have a dendrogram with two leaves, and we want to apply a function f to each node a,b,c.
Tree:
a
/ \
b c
Initialize LL:
HEAD -> NULL
Add root node:
HEAD -> a -> NULL
Set ptr to head:
HEAD -> a* -> NULL
Iterate over linked list:
HEAD -> a* -> c -> NULL (insert child, back to front to ensure correct traversal)
HEAD -> a* -> b -> c -> NULL (insert child)
HEAD -> a -> b* -> c -> NULL (increment pointer)
HEAD -> a -> B* -> c -> NULL (b has no children; apply f, f(b)=B)
HEAD -> aB* -> c -> NULL (merge B into parent)
HEAD -> aB -> c* -> NULL (a has an unevaluated child, so increment pointer)
HEAD -> aB -> C* -> NULL (apply f to c, f(c) = C)
HEAD -> aBC* -> NULL (merge C into parent)
HEAD -> ABC* -> NULL (a has no unevaluated children so apply f, f(a)=A)
HEAD -> ABC -> NULL* ( done, return ABC )
This implementation achieves the same result as a recursive operation, but doesn’t run the risk of overflowing the stack for deep trees. Note that this algorithm is a post-order traversal; pre-order is the default and is discussed later.
Details and Difficulties
There are a few things about the implementation of this that make things difficult. First, the R garbage collector likes to collect R objects while they’re being worked on in C. We can get around this by using PROTECT() to tell the garbage collector not to touch the object, but the protection stack has a fixed size that is extremely small (10,000 items by default). The tree as a whole can be protected when it’s first loaded, but calling f() on a node of the tree produces a new R object that is unprotected, so we have to protect the result.
The naive approach is to just put all the nodes into a linked list, apply the function to every node, then rebuild the tree. Unfortunately, this rapidly exhausts the available space we have on the protection stack. Instead, we have to work slightly smarter.
We can get around this issue by immediately assigning the value of f(node) to the parent node. If the parent node is protected, the value assigned to it will be protected as well. This means that, as long as we ensure the parent is always protected, we don’t need any extraneous protection calls. Some clever shuffling can ensure that we protect everything under the first PROTECT call on the entire tree.
I’ve implemented two methods for dendrapply: a pre-order traversal and a post-order traversal. The final implementation for both uses a maximum of 3 slots on the protection stack, and no recursive function calls. The original tree consumes the first PROTECT call, which protects all its children until they’re modified. When each node is evaluated, we use a PROTECT call to create the R expression to be called and a second to protect the SEXP returned from the function. This value is then assigned to the parent node using SET_VECTOR_ELT, which protects the value. Since protection is by value and not by reference, we can safely store this new value in the linked list. Applying the function to the root can safely be done by using REPROTECT() to preserve protection on children.
The result is a little funky in the pre-order case: we use VECTOR_ELT on the parent to get the node, call the function on it, replace the node using SET_VECTOR_ELT on the parent, and then populate the reference in the linked list by calling VECTOR_ELT on the parent a second time. This ensures we always have protected values.
With a post-order traversal, the implementation is a little cleaner. Since the children of each node are always evaluated prior to the node itself, we don’t need to recall VECTOR_ELT after calling the function. Instead, we can just merge the nodes and continue.
The main difference for end users between the two traversals is that, for a given node n, pre-order traversal will always evaluate f(n) before any of its children, whereas post-order traversal will always evaluate f(n) after all of its children. Pre-order is the default for backwards compatibility with the original stats::dendrapply, which used a pre-order traversal. The post-order method allows for some new functions, such as the following:
f <- function(x){
if(!is.null(attr(x, 'leaf'))){
v <- as.character(attr(x, 'label'))
} else {
v <- paste0(attr(x[[1]], 'newattr'), attr(x[[2]], 'newattr'))
}
attr(x, 'newattr') <- v
x
}This function assigns a new attribute equal to the label if it’s a leaf, or the concatenation of the child nodes’ new attributes if it’s an internal node. The default application of dendrapply will only create new attributes for the leaves, and will return character(0) for any internal nodes (since the children won’t have had their new attribute set yet). However, using how='post.order' will ensure we evaluate the children first, meaning that internal nodes will be assigned a non-empty value:
# dendrogram with 3 leaves and two internal nodes
dend <- as.dendrogram(hclust(dist(1:3)))
# original dendrapply from `stats`
stats::dendrapply(dend, exFunc)
attr(dend, 'newattr')
# > character(0)
# pre-order (default)
dendrapply(dend, exFunc, how='pre.order')
attr(dend, 'newattr')
# > character(0)
# post-order
new_dendrapply(dend, exFunc, how='post.order')
attr(dend, 'newattr')
# > "312"This capability is something I have found myself wishing for often in dendrapply. Calculating Fitch Parsimony for a phylogeny is a great example of a method that relies upon a post-order traversal.
Replicating Bugs
Another weird thing I had to contend with is ensuring that the functionality exactly replicates that of the current dendrapply so as not to break existing packages. I’m reminded of this tweet:

Users tend to do things with code the developers never expect, which is exactly how I found myself failing all build checks repeatedly when trying to integrate my code into my fork of the R codebase. It turns out, at one point, a test was added to ensure dendrapply performs expectedly when doing the following operation:
> D <- as.dendrogram(hclust(dist(cbind(setNames(c(0,1,4), LETTERS[1:3])))))
> dendrapply(D, labels)
[[1]]
[1] "C"
[[2]]
[[2]][[1]]
[1] "A"
[[2]][[2]]
[1] "B"
[[3]]
[1] "C"
“Expectedly” means producing the output shown. This was surprising to me, because this dendrogram is fully bifurcating. Additionally, labels(D) produces c("C", "A", "B"), so it’s not even that we performed labels() on the root node and then replaced the first and second entries with the result of their children. It turns out that, due to the way the original dendrapply was coded, the results of each operation replicate if they’re not large enough to fill the list generated by the parent. This means that, after applying labels() to the root node, we get a list with 3 elements. Then, when we apply labels to the children, we get two new values. The parent list has three values, so we replicate those values as many times as necessary. In this case, the result of the children are the first two entries shown above. Since the list has length 3, we replicate the first element to the third location, producing the output given.
According to the tests, many CRAN packages rely on this bug working as “expected”. This required a bit of a code refactor, but I did manage to get my C code to replicate values across the parent array to accurately reproduce this bug. Knowing how software development is, I’m sure that I’ll have to do something similar for future bugs people are relying on…but that’s an issue for another day.
Tentative Future Additions
A inorder traversal is definitely possible, but I’m not sure if it’s worth it. The implementation would use the almost same code as the post-order case, although nodes would be added by inserting the right element next and the left element at the end of the list. In-order traversal on a multifurcating tree is defined as evaluating all but the last child node, then the node, then the final (rightmost) child. I can’t think of a good use case for this type of traversal, especially for multifurcating trees.
Breadth-first traversals are also easily implementable by making elements insert at the end of the linked list rather than at the next position. However, the behavior of these can be a little odd with dendrogram objects (the result is fairly counterintuitive to me at least). These may be worth exploring as options in the future.
Something I would like to implement is a “flat” application of dendrapply, similar to the flexibility offered in rapply. Providing an option to get the results as a flat list/vector could have very good usecases. To illustrate what this would look like, imagine the following tree with shown labels:
a
/ \
b c
/ \ / \
e f g h
I’d like the function to be able to do something like:
> dendrapply(exTree, \(x) attr(x, 'label'), how='post.order', flatten=TRUE)
[1] "e" "f" "b" "g" "h" "c" "a"
> dendrapply(exTree, \(x) attr(x, 'label'), how='pre.order', flatten=TRUE)
[1] "a" "b" "e" "f" "c" "g" "h"
Note that this is different from dendrapply in that the result is a flat vector and not a nested list, and different from rapply in that the result is the function applied to leaves and internal nodes. Adding breadth-first and in-order traversals would likely be more useful for this kind of function than for the standard dendrapply.
Benchmarking
Speed gains from this implementation are relatively modest, although I suspect that further optimization could improve runtime. As the main improvement of this is in the backend and not the R function calls themselves, it should have relatively consistent performance regardless of the input function. Testing was performed on a simple function to add an attribute to each node, as well as a recursive one that calls rapply at every node. Speedup is relatively consistent regardless of function, with the average boost in runtime approximately 1.5-3x on my machine (2021 MacBook Pro, M1 Pro, 32GB RAM). Calling rapply had less of a speedup compared to faster functions, likely because the runtime of the called R function dominates the overall runtime of the dendrapply call. Benchmarking using a minimal function to measure only the impact of the new apply function resulted in an average speedup of 2.5-3x depending on tree size. Looking at the memory usage of the functions, my new implementation has significantly decreased usage due to fewer function call frames allocated on the stack. However, this is difficult to benchmark since the original function uses R to allocate memory and the new function allocates in C.
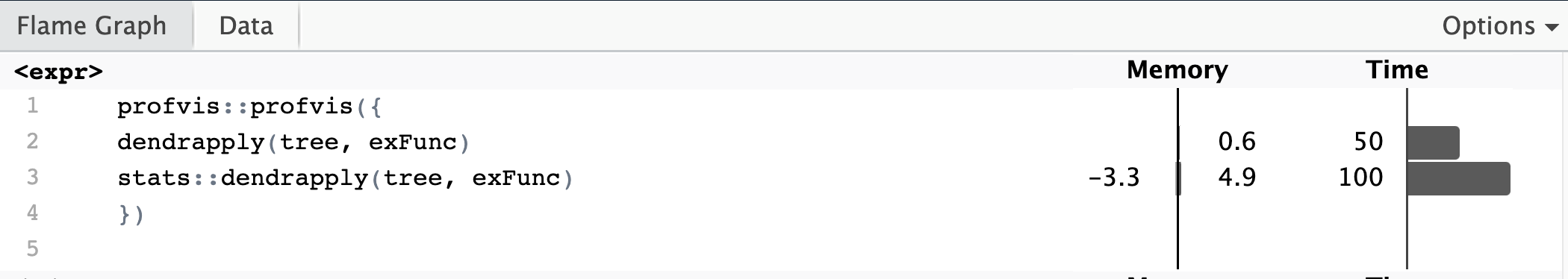
Compatibility with the previous version of dendrapply was tested against the unit tests available in dendextend, one of the largest packages that makes extensive usage of stats::dendrapply in a variety of scenarios. The pre-order traversal version passed all unit tests.
Post-order failed many tests because methods in dendextend depend on the pre-order traversal order–this is why pre-order is the default setting.
Complete Code:
The complete code is available on Github. What follows are some comments on the code contained within the files.
R Script
This has some quirks to make it a drop-in replacement for stats::dendrapply. The behavior of the original function when the provided function does not return a dendrogram or list-like object is a little counterintuitive to me, but after lots of testing this implementation should replicate it all accurately. There is a weird quirk where calling VECTOR_ELT on an SEXP seems to unclass the object, and I was running into problems with the input nodes not being of type dendrogram. I thought it was fairly safe to just reclass the object as 'dendrogram' when it comes to the function, since we expect to be applying the function to dendrogram objects anyway (and children of a dendrogram should also be dendrogram).
C Code
The C code is much longer. The main functions exposed to R are do_dendrapply() via .Call interface, and free_dendrapply_list() via .C interface. The brunt of the computation is done in new_apply_dend_func, which will likely be either renamed in the future or rolled into do_dendrapply(). Note that checking for leaf nodes depends on the leaves having correct values for attr(node, 'leaf'); if someone messes with the nodes it could perform unexpectedly. The current implementation defines leaves as nodes such that attr(node, 'leaf')==TRUE and non-leaves as is.null(attr(node, 'leaf')) || attr(node,'leaf')==FALSE. It’s unfortunately not possible to just check for nodes with length 1, since we should be able to support dendrograms with arbitrary numbers of children at internal nodes (whether that’s 1, 2, or many). I regard this error as being in the same category as users being able to change S3 classes to arbitrary values–it’s known and documented that things could go wrong if users mess with the values, and those that choose to modify the values are proceeding at their own risk. I’m not completely sure what the function will do in this scenario–I believe it will probably run infinitely until the C memory limit is exceeded. I’ve added enough checks to ensure that the function execution is always rescuable via user interrupts, so I think this is overall okay.
