Automated R Testing and Coverage on GitHub (without Codecov!)
Published:
I’ve recently been working on adding some unit testing to Biostrings as part of working I’m doing for a grant awarded by R’s Infrastructure Steering Committee. As part of implementing unit tests, I wanted to add an automatic workflow that would evaluate the current tests on any future pull requests, so that (1) we could be sure contributed code isn’t breaking any additional functionality (2) we can be sure that contributed code is adequately tested.
The easiest way to do this is to set up testing with testthat and covr, and then use Codecov.io to store coverage reports. There’s even a GitHub action already set up to do this! My first attempt was to do exactly that–testing and code coverage is calculated with this GitHub action, results are uploaded to Codecov, and Codecov automatically adds a comment to the PR with the status.
Unfortunately, it turns out that this isn’t super feasible for Biostrings. Codecov is free for single users, but begins to get much more expensive as you scale up to more and more users. Additionally, Codecov needs access to at least one user’s OAuth token, and they have to be an admin of the repository in question to be able to add comments to PRs. As I am both broke and not the owner of the Biostrings repository, this unfortunately isn’t a feasible solution.
My solution: a custom workflow
There are three things I wanted to accomplish with my workflow:
- Run all unit tests in the package, and report any that had unexpected behavior
- Calculate the code coverage of tests, and how its changed relative to the base repository
- Report results to users in a way that doesn’t require elevated permissions
I ended up writing my own workflow. It works for R packages using testthat as a testing environment, but could easily be adapted to any other testing setup. The result is printed on the Job Summary page, and renders into something like this:
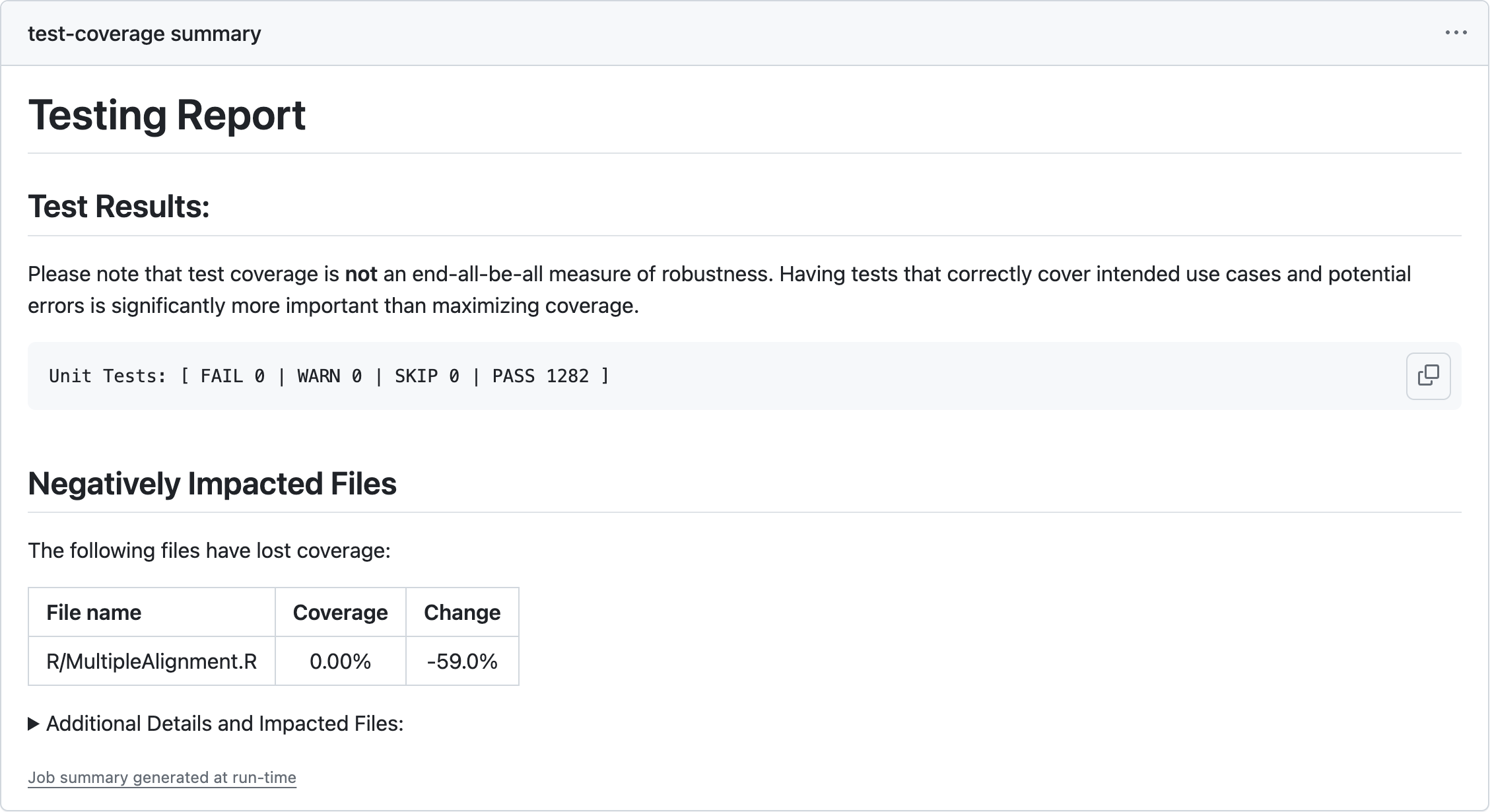
Main output
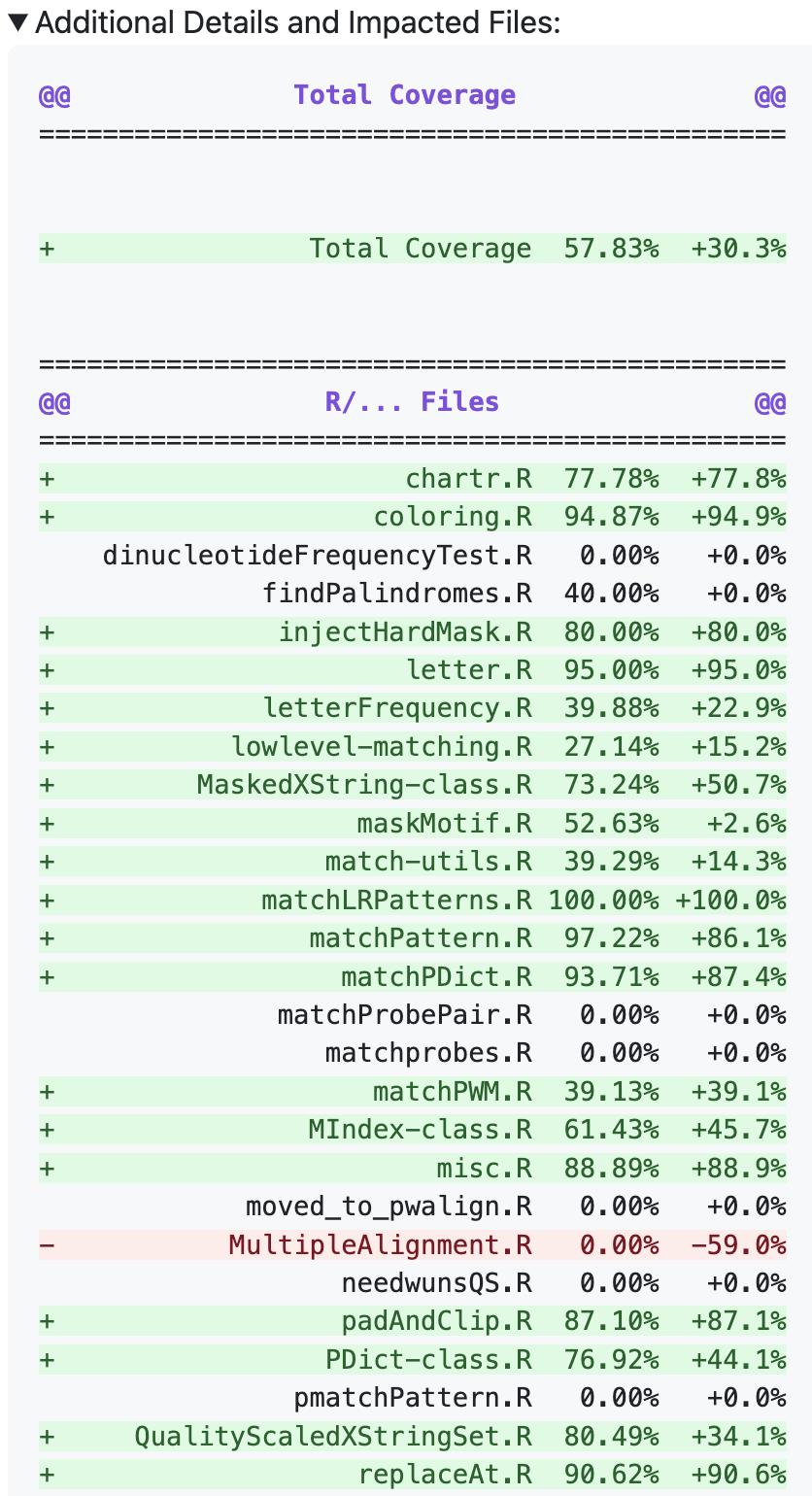
Expanded table
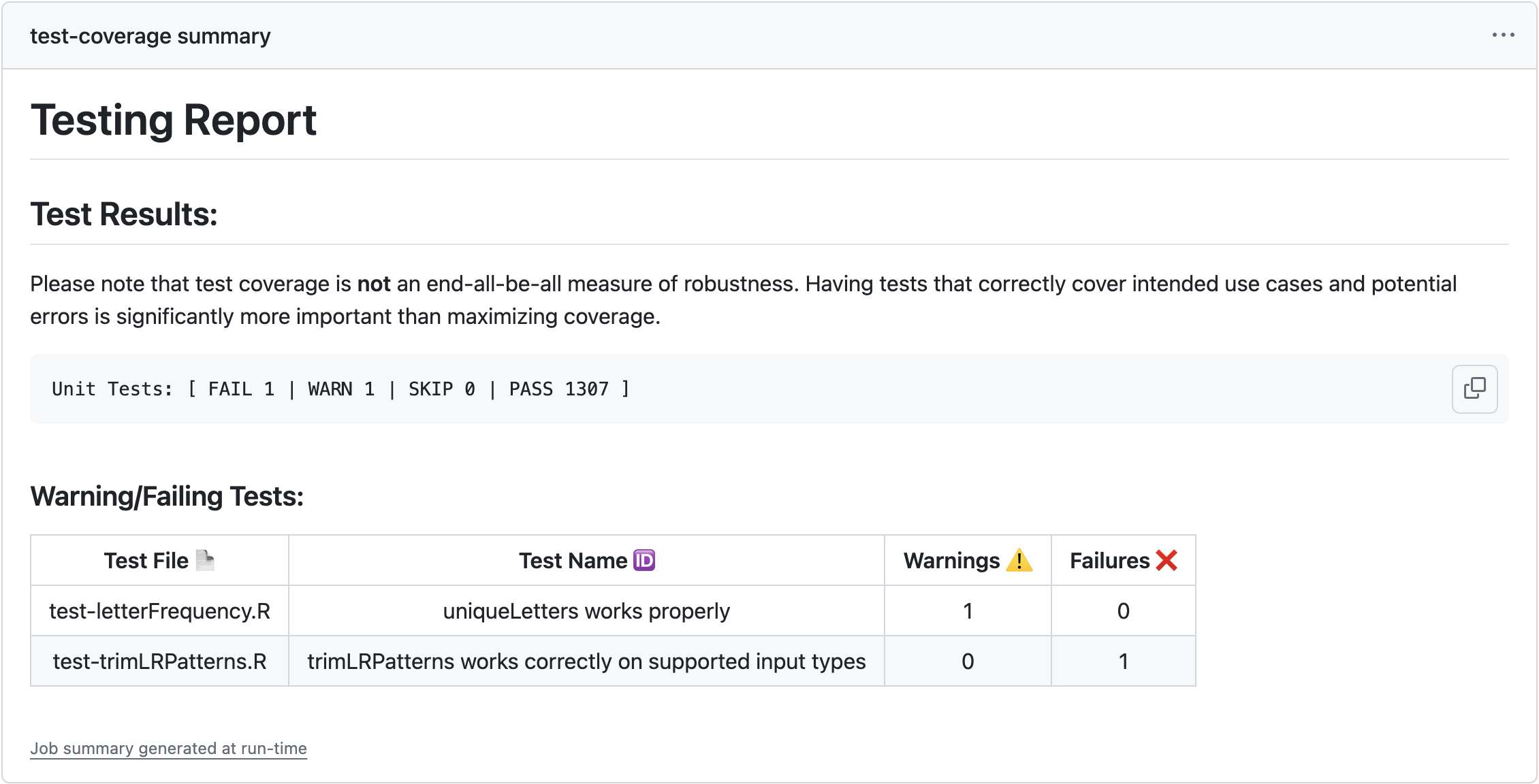
Some failing tests
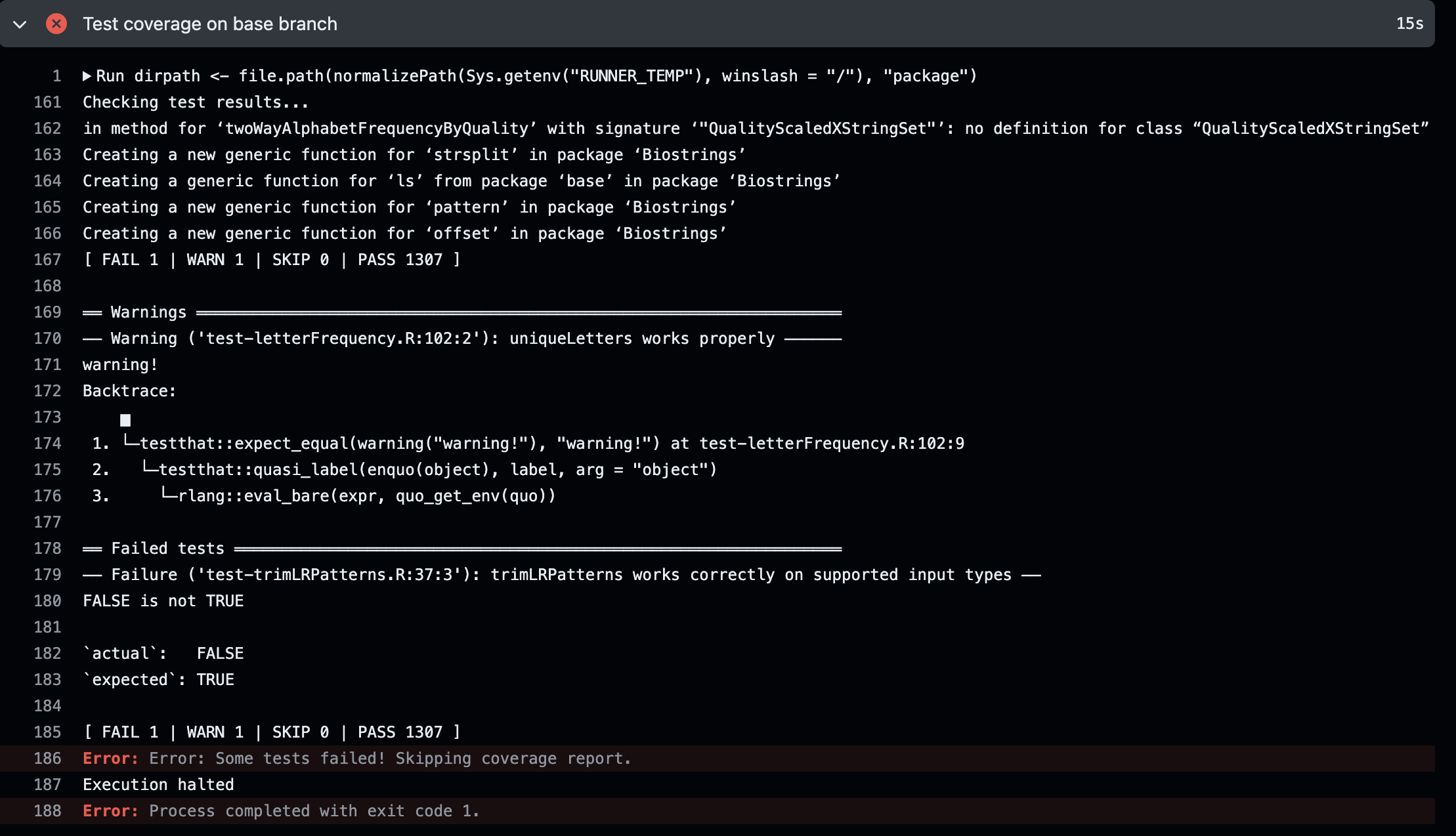
Failing test detail
How does it work?
The process essentially looks like this:
- Pull the code in the PR
- Pull the base repository code (the current version)
- Install R and set up dependencies for both packages
- Check unit tests on the new code
- Calculate code coverage on new code and the base repository, and report changes
- Display all results to users
Steps 1-3 are fairly easy using prebuilt GitHub Actions. We can do all of them with a few lines:
steps:
- name: Checkout current ref
uses: actions/checkout@v4
with:
path: ./new-state
- name: Checkout Biostrings devel ref
id: devel-checkout
uses: actions/checkout@v4
with:
repository: Bioconductor/Biostrings
path: ./original-state
- uses: r-lib/actions/setup-r@v2
with:
use-public-rspm: true
- name: setup dependencies for old state
uses: r-lib/actions/setup-r-dependencies@v2
with:
working-directory: ./original-state
extra-packages: any::covr
needs: coverage
- name: setup dependencies for new state
uses: r-lib/actions/setup-r-dependencies@v2
with:
working-directory: ./new-state
extra-packages: any::covr
needs: coverage
This loads the contributed code into ./new-state and the original repo’s base branch code into ./old-state.
After this, I call a long R script that accomplishes (4-5). The script does the following:
First, we run the unit tests on the new version of the package:
## first check unit tests
cat("Checking test results...\n")
res <- testthat::test_local("./new-state", stop_on_failure=FALSE, reporter="check")
res <- as.data.frame(res)
test_report <- c(sum(res$failed), sum(res$warning), sum(res$skipped), sum(res$passed))
Throughout this process, I’m going to build an object called out_msg. This variable holds a Markdown-formatted string that we’ll eventually print as output to users. At this point, I’m going to initialize out_msg, and then add in some diagnostic information on tests that failed or threw warnings (if any):
## build the output message
out_msg <- '# Testing Report\n\n'
out_msg <- paste0(out_msg, "## Test Results:\n\n")
out_msg <- paste0(out_msg, "Please note that test coverage is **not** an ",
"end-all-be-all measure of robustness. Having tests that ",
"correctly cover intended use cases and potential errors ",
"is significantly more important than maximizing coverage.\n\n")
out_msg <- paste0(out_msg, "```\n", test_report, '\n```\n\n')
## if any tests failed or threw warnings, report them
shouldStop <- test_report[1] > 0
shouldPrint <- sum(test_report[1:2]) > 0
test_report <- paste(c("FAIL", "WARN", "SKIP", "PASS"), test_report, collapse=' | ')
test_report <- paste('Unit Tests: [', test_report, ']')
if(shouldPrint){
## put failing/warning tests into a markdown table
p_toprint <- which(res$failed + res$warning > 0)
ptp <- res[p_toprint,]
failed_tests <- ptp[,c("file", "test", "warning", "failed")]
failed_tests <- apply(failed_tests, 1L, paste, collapse=" | ")
failed_tests <- paste("|", failed_tests, "| ")
failed_tests <- paste(failed_tests, collapse='\n')
md_tab <- paste0("| Test File :page_facing_up: | Test Name :id: | Warnings :warning: | Failures :x: | \n",
"| :-----: | :-----: | :-----: | :-----: | \n",
failed_tests, "\n\n")
out_msg <- paste0(out_msg, "### Warning/Failing Tests:\n\n", md_tab)
}
if(shouldStop){
## if any errored, stop the workflow without continuing to test coverage
cat(out_msg, file='./test_status.md')
stop("Some tests failed! Skipping coverage report.")
}
Notice that if any tests failed, we stop early. This is because there isn’t a good way to run covr with tests that fail. testthat allows for test to continue running even if some fail, whereas covr doesn’t support that option. Additionally, if a test failed, fixing that is more important than calculating coverage anyway.
Next, we calculate the coverage on both the old and new code. This is done on every run–it’s probably possible to cache results on the base repository, but it adds some complexity and wasn’t a big enough improvement in performance for me to want to implement it.
## if no tests failed, check coverage of old vs. new
library(covr)
# exclude lines with no content
options(covr.exclude_pattern=c("^[ \t{}()]+$"))
# get results on old state
files_to_ignore <- list("R/AMINO_ACID_CODE.R", "R/GENETIC_CODE.R",
"R/zzz.R", "R/IUPAC_CODE_MAP.R",
"R/getSeq.R")
## old state coverage
cov <- covr::package_coverage(
path = "./original-state",
quiet = FALSE,
clean = FALSE,
install_path = file.path(dirpath, "old-state"),
function_exclusions = "^\\.",
line_exclusions=files_to_ignore
)
head_res <- covr::coverage_to_list(cov)
## new state coverage
cov <- covr::package_coverage(
path = "./new-state",
quiet = FALSE,
clean = FALSE,
install_path = file.path(dirpath, "new-state"),
function_exclusions = "^\\.", # excludes functions starting with .
line_exclusions=files_to_ignore
)
new_res <- covr::coverage_to_list(cov)
## compare difference in coverage
f_old <- head_res$filecoverage
f_new <- new_res$filecoverage
Now that we have coverage on both the old and new code versions, a lot of code goes into transforming raw values into pretty printing for the eventual Markdown file.
## get changes in coverage for each file
all_files <- union(names(f_old), names(f_new))
file_changes <- rep(0, length(all_files))
names(file_changes) <- all_files
file_changes[names(f_new)] <- file_changes[names(f_new)] + f_new
final_cov <- file_changes ## this is the new coverage of all files
file_changes[names(f_old)] <- file_changes[names(f_old)] - f_old
## get the change in overall coverage
total_change <- new_res$totalcoverage - head_res$totalcoverage
## start appending output information
out_msg <- paste0(out_msg, "## Negatively Impacted Files\n\n")
## If any files lost coverage, add them here
n <- names(file_changes)
pos_neg <- which(file_changes < 0)
if(length(pos_neg) > 0){
pos_neg <- pos_neg[order(file_changes[pos_neg], decreasing=FALSE)]
warn_changes <- sprintf("%+.01f%%", file_changes)
header <- "| File name | Coverage | Change |\n | :----- | :-----: | :-----: |\n"
warn_tab <- paste0('| ', n[pos_neg], ' | ', sprintf("%0.02f%%", final_cov[pos_neg]), ' | ',
unname(warn_changes[pos_neg]), ' |', collapse='\n')
warn_tab <- paste0(header, warn_tab)
out_msg <- paste0(out_msg, "The following files have lost coverage:\n", warn_tab, '\n')
} else {
out_msg <- paste0(out_msg, "No negatively impacted files. Nice job!\n\n")
}
I’ll note for this next part that using diff-formatted Markdown is very nice for displaying file changes. You can tag lines as + to highlight them in green, and tag them as - to highlight them in red. All these lines put the file-specific coverage and changes in coverage into a diff-formatted table for future printing. We also enclose it in a <details> block so that it can be hidden, since this is a lot of extra content.
## build extended diff table
## going to split the files by whether or not they're R files or src files
p_Rfiles <- grepl("^R/", n)
## lots of formatting to get consistent entry widths for the diff table
n <- vapply(strsplit(n, '/'), .subset, character(1L), 2L)
all_diffs <- data.frame(filename=n,
coverage=sprintf("%.02f%%", final_cov),
change=sprintf("%+.01f%%", file_changes))
max_nchar <- max(nchar(all_diffs$filename))
all_diffs$filename <- sprintf(paste0("%", max_nchar, "s"), all_diffs$filename)
all_diffs$coverage <- sprintf("%7s", all_diffs$coverage)
all_diffs$change <- sprintf("%7s", all_diffs$change)
all_diffs$mark_char <- 1L
all_diffs$mark_char[file_changes > 0] <- 2L
all_diffs$mark_char[file_changes < 0] <- 3L
all_diffs$mark_char <- c(" ", "+", "-")[all_diffs$mark_char]
all_rows <- apply(all_diffs[c(4,1:3)], 1L, paste, collapse=' ')
w <- nchar(all_rows[1L])
## multiple sections, each title has to be centered within its block
## Section 0: total coverage
title0 <- "Total Coverage"
n_padding <- (w - nchar(title0) - 4) / 2
title0 <- paste0("@@", paste(rep(' ', floor(n_padding)), collapse=''),
title0, paste(rep(' ', ceiling(n_padding)), collapse=''), "@@")
row0 <- paste(ifelse(total_change < 0, "-", ifelse(total_change>0, "+", " ")),
sprintf(paste0("%", max_nchar, "s"), "Total Coverage"),
sprintf("%6.02f%%", new_res$totalcoverage),
sprintf("%+6.01f%%", total_change), collapse=' ')
## Section 1: R file coverage
title1 <- "R/... Files"
n_padding <- (w - nchar(title1) - 4) / 2
title1 <- paste0("@@", paste(rep(' ', floor(n_padding)), collapse=''),
title1, paste(rep(' ', ceiling(n_padding)), collapse=''), "@@")
## Section 2: src file coverage
title2 <- "src/... Files"
n_padding <- (w - nchar(title2) - 4) / 2
title2 <- paste0("@@", paste(rep(' ', floor(n_padding)), collapse=''),
title2, paste(rep(' ', ceiling(n_padding)), collapse=''), "@@")
## section dividers
spacer <- paste(rep('=', w), collapse='')
entries1 <- paste(all_rows[p_Rfiles], collapse='\n')
entries2 <- paste(all_rows[!p_Rfiles], collapse='\n')
## final table
diff_table <- paste(title0, spacer, '\n', row0, '\n', spacer,
title1, spacer, entries1, spacer,
title2, spacer, entries2, spacer,
collapse='\n', sep='\n')
## enclose it in a <details> tag so it doesn't take up a ton of space
diff_table <- paste0("<details>\n<summary>Additional Details and Impacted Files:</summary>\n\n",
"```diff\n", diff_table, '\n\n```\n\n</details>')
out_msg <- paste0(out_msg, diff_table, '\n')
cat(out_msg, file='./test_status.md')
All of that content is put into a file called test_status.md. At first, I was using a GitHub Action to attach this file as a comment in the PR, but that unfortunately requires elevated permissions in the GitHub token used to run it. Even worse, it isn’t possible to auto-generate a token with the necessary permission pull-request: write on public repositories (for security concerns), so I’d have to ask Bioconductor to create me a token to use. Given that GitHub thinks this is a security risk, I decided I’d rather not risk it and go through the hassle.
It turns out that there’s another option. GitHub Actions have recently release Job Summaries, which allow you to print information about your workflow steps without requiring any additional steps. It unfortunately can’t print to PRs (yet), but it’s close enough and requires no elevated permissions. The remainder of my workflow file just needs to call the following steps:
- name: Print results to summary
if: always()
run: cat ./test_status.md >> $GITHUB_STEP_SUMMARY
- name: Upload status on success
if: always()
uses: actions/upload-artifact@v4
with:
name: coverage-test-results
path: ./test_status.md
The first of these prints the contents of test_status.md to the Job Summary page, and the second uploads the raw markdown file. The if: always() tag ensures that this runs even if jobs fail, meaning that if we abort early (due to a failing test, for example) we still get output.
Conclusion
If you’re interested in the workflow, you can check it out on GitHub. Try it out for yourself if you’d like! In the future, I’m going to add more workflows to automatically run R CMD CHECK and BiocCheck::check() so that we can be sure the package is always ready to be pushed to Bioconductor. For other updates, follow the progress of my grant work here!
